Systems Immunology Research in the Dolatshahi Lab
Double vision: Seeing lung cancer by eye and AI, Image courtesy of Remziye & Gabe
Systems Immunology Research in the Dolatshahi Lab
The immune system is complex and ubiquitous. Numerous immune cell types either reside in a particular tissue or circulate throughout the body. Each cell type has its unique roles and approaches in sensing problems, communicating with other cells, and eliciting their immune functions. The Dolatshahi lab takes a systems approach to understand the interconnecting pathways that control immune responses spanning across multiple biological systems and scales.
This understanding will enable us to optimize immune responses to confront specific issues, ranging from infections to cancer. By building computational models and machine-learning strategies that integrate experimental data across various molecular and cellular scales, Dolatshahi laboratory seeks to: (1) ascertain how biophysical properties of the immune factors (defined as their subclass and post-translational modifications) determine their function, (2) uncover mechanisms responsible for dysregulation of these properties in disease states, and (3) use this knowledge to guide biomarkers for early disease diagnosis, patient stratification and optimized immunotherapies. Another key step in translational immunotherapies is the design of maximally informative animal studies for reliable prediction of the outcome in humans. To this end, Dolatshahi lab also (4) creates predictive computational transfer functions to relate mechanistic models of cellular processes in animals to those in humans. Such functions will enable inter-species translation of molecules to phenotypes.
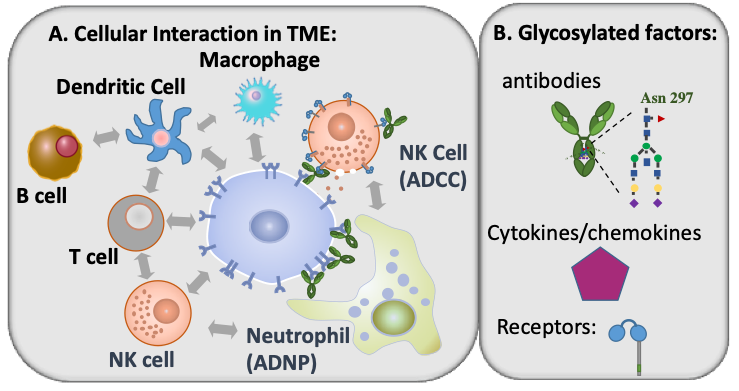
Systems Biology of Immune Factor Glycosylation and Intercellular Interactions in the Tumor Microenvironment
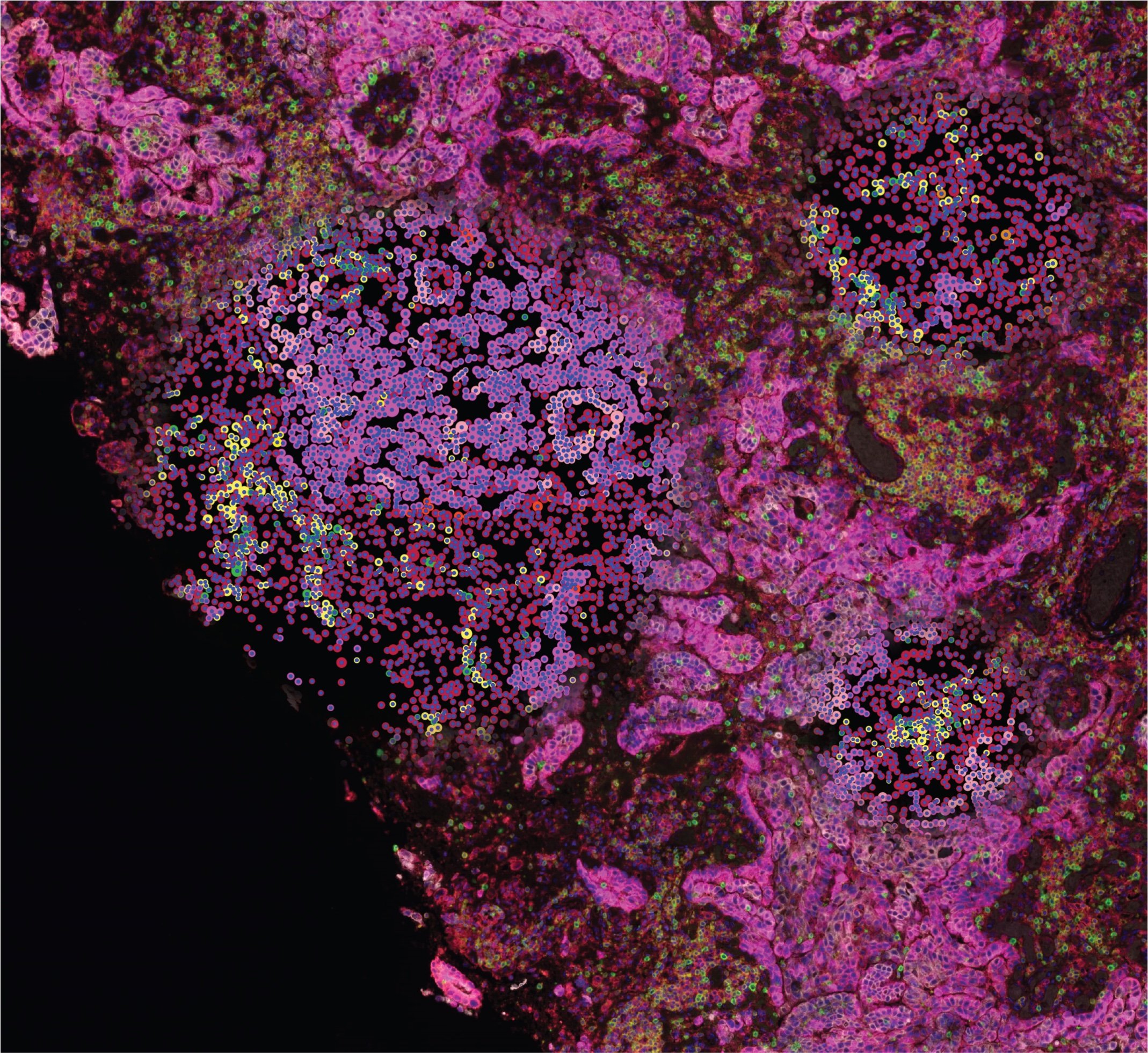
The role of micro-environmental interactions between tumor, stromal and immune cells have been extensively demonstrated in cancer growth, tissue remodeling, angiogenesis, migration, and tissue invasion. Tumor-inhibiting and tumor-promoting tumor-immune interactions have been identified as prognostic markers and immunotherapeutic targets such as checkpoint inhibitors. The prognostic and therapeutic success of these solutions, however, have been limited likely due to the highly complex stromal-tumor-immune interactions present in the tumor microenvironment (TME). The cellular interactions in the TME are mediated by ligand-receptor interactions, the complexities of which are further multiplied by the diverse post-translational modifications (PTM) of these secreted and cell-surface proteins. Glycosylation is one of the most common types of PTM and a critical determinant of the function of proteins, namely immune factors, such as immunoglobulins (Ig), tyrosine kinase receptors, integrins, and several other receptors and cytokines. Alterations in glycosylation are common in various cancer contexts and vary by disease progression. While protein glycosylation and TME alterations are known cancer hallmarks, a combined systems analysis of these inter-related changes is lacking. Accordingly, the overall goal of this project is to identify novel tumor-inhibiting or tumor-promoting interactions that are influenced by or impact glycosylation. These glycan-mediated interactions present novel markers for improved prognosis and introduce novel immunotherapeutic targets.
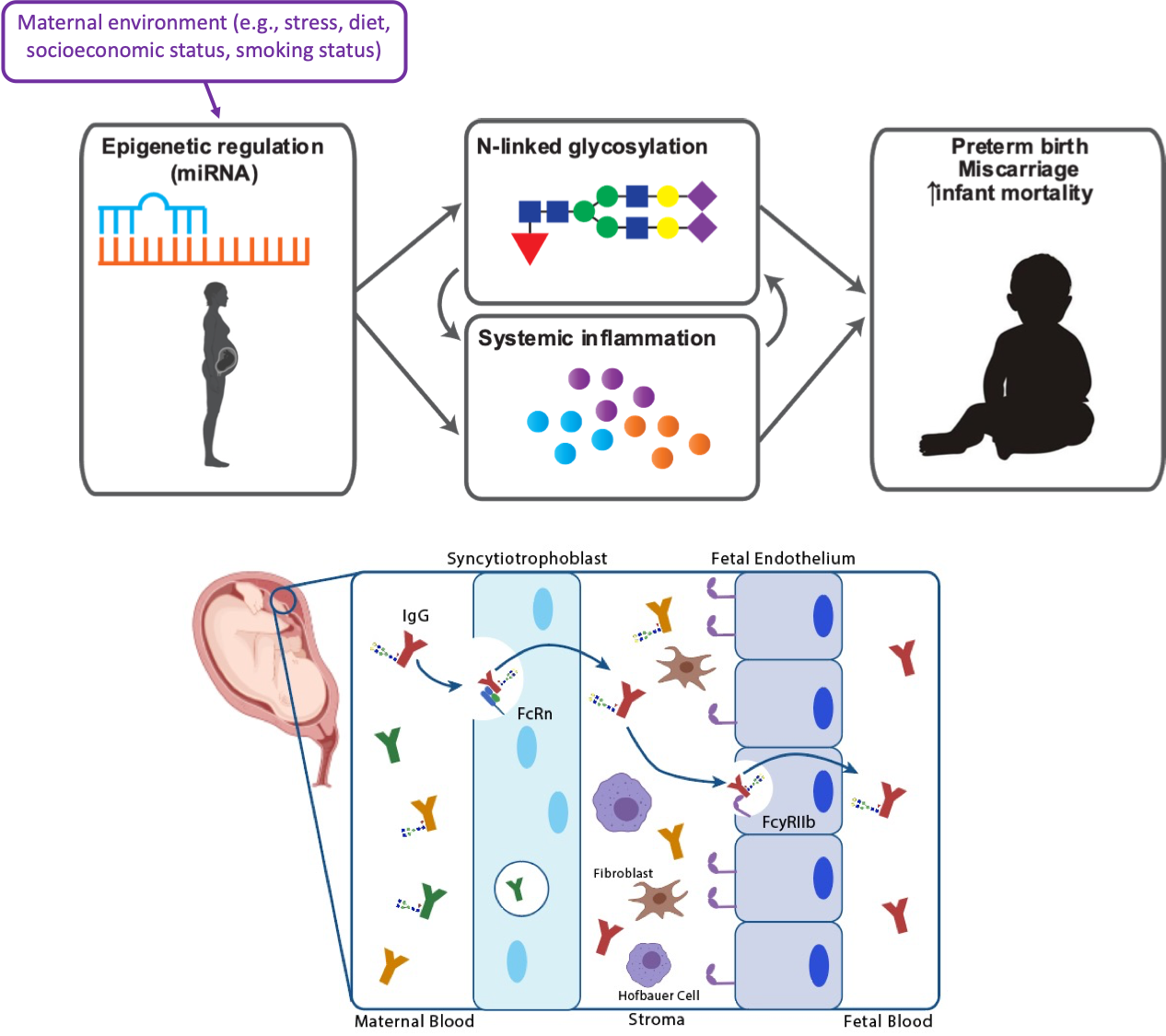
Using systems immunology to understand immune reprogramming during pregnancy and the transplacental transfer of antibodies from mother to child
Systems serology attempts to quantify the entire humoral response via functional and biophysical assays. In a collaborative systems serology project, we set out to define the rules that govern transfer of antibody functionality across placenta from mother to child. We examined paired maternal and cord blood of a cohort of mothers and children for their antigen-specific immune profiles including: (a) antibody subclasses, (b) glycan profiles and the incurred antibody dependent phagocytosis by neutrophils and monocytes, (c) cytotoxicity and secretion of cytokines and chemokines by natural killer (NK) cells, and (d) complement deposition. Using Partial Least Squares Discriminant Analysis (PLSDA), we first defined the top contributors to the immune profile differences between moms and infants, including NK cell functions, IgG1 and doubly-galactosylated IgG. Next, we developed a statistical modeling framework that sets robust criteria to determine the active transfer, blocking or abundance-strict transfer of antibody attributes.
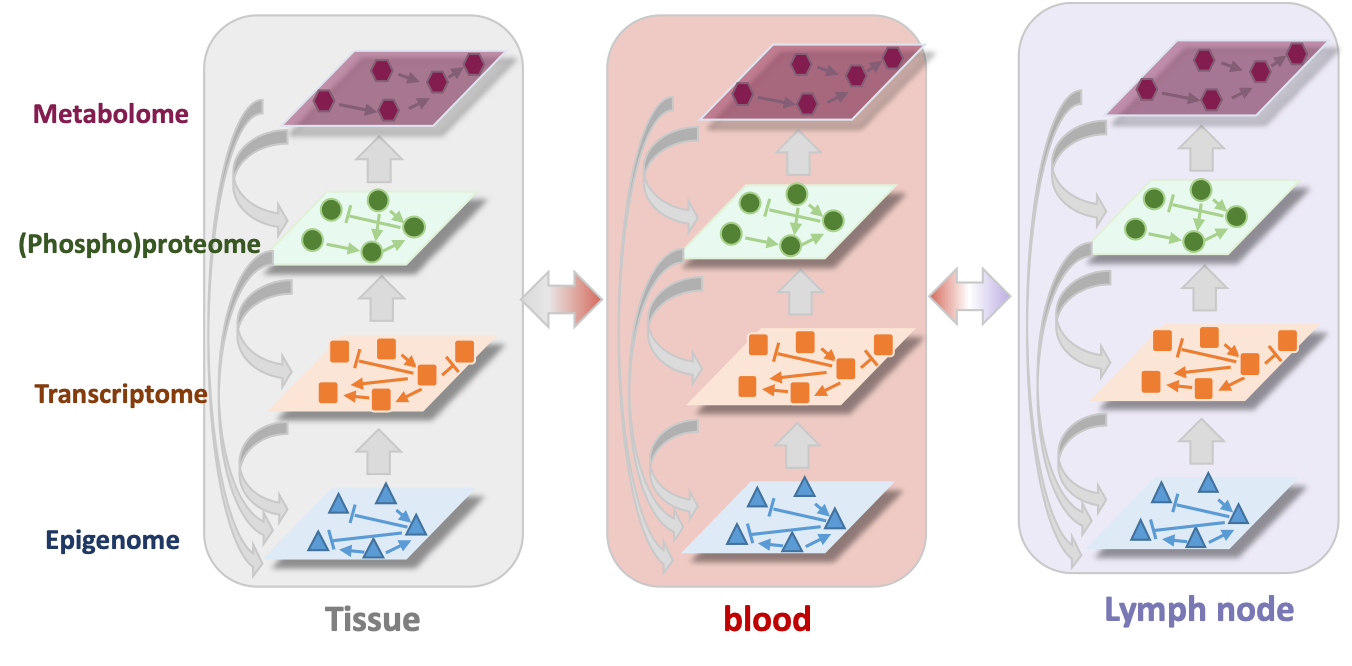
Computational Biology – Multi-scale modeling of molecular and cellular networks
Emerging technologies to profile complex biological phenomena increasingly drive advances in biomedical sciences. These technologies provide multi-modal snapshots of biological systems at a range of spatial and temporal scales. To weave mechanistic stories based on a multitude of intracellular molecular measurements, inter-cellular, tissue-level and functional data, sophisticated computational methods are required. To this end, we combine multiplex experimental measurements with computational methods to solve problems in the context of cancer, infectious disease and maternal-neonatal immunology among others. A central paradigm of our research is development and testing of integrative computational models including data-driven statistical machine learning, network inference, information theory, and signal processing as well as bottom-up mechanistic kinetic-dynamic modeling.
Our research is made possible with funding support from these organizations:
ACS IRG